
Searching for microbial antibiotic resistance in urban stormwater
Why do we look for antibiotic resistance?
In D4RUNOFF we measure the levels of microbial resistance to antibiotics in urban stormwater and there are good reasons for this. According to the World Health Organization: “Antimicrobial resistance in infectious agents represents a global health security threat and continues to be a serious threat to human, animal and environmental health, as well as the well-being of the global economy” (WHO, 2022). Urban stormwater certainly is not the major exposure route for antibiotic resistant bacteria, but we suspect that some types of stormwater may be an overlooked reservoir of microbial antibiotic resistance. When designing nature-based solutions for local handling of stormwater, it is therefore important to know the levels, sources, and fate of antibiotic resistance, so that the solutions can be designed to reduce exposure to antibiotic resistant bacteria.

How can we measure antibiotic resistance?
We have two approaches for measuring antibiotic resistance, one is based on classical growth of antibiotic resistant bacteria on selective medium, the other is based on direct quantification of selected antibiotic resistance genes. When we get a stormwater sample, we first test for growth of antibiotic resistant enterobacteria. This is a method where the traditional agar plate is substituted with simple petrifilms that do not need to be sterilized and may be used in the field. Only gut bacteria from the group Enterobacterales can grow on this type of petrifilms and we have added either amoxicillin, tetracycline, ciprofloxacin or trimethoprim, so only enterobacteria resistant to these common antibiotics will grow. After 24 hours growth, we can simply count the number of resistant enterobacteria colonies on the petrifilms. As controls, we take colonies from the petrifilms and grow them on traditional agar-plates with antibiotics followed by DNA-sequencing to identify the bacteria. Fortunately, the petrifilm colonies have turned out to be both antibiotic resistant and enterobacteria as expected.
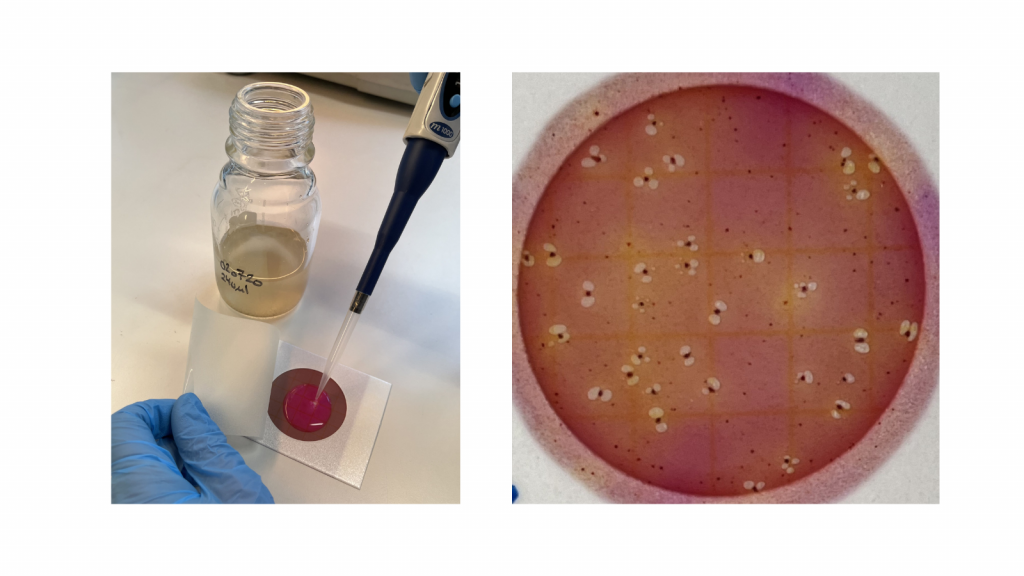
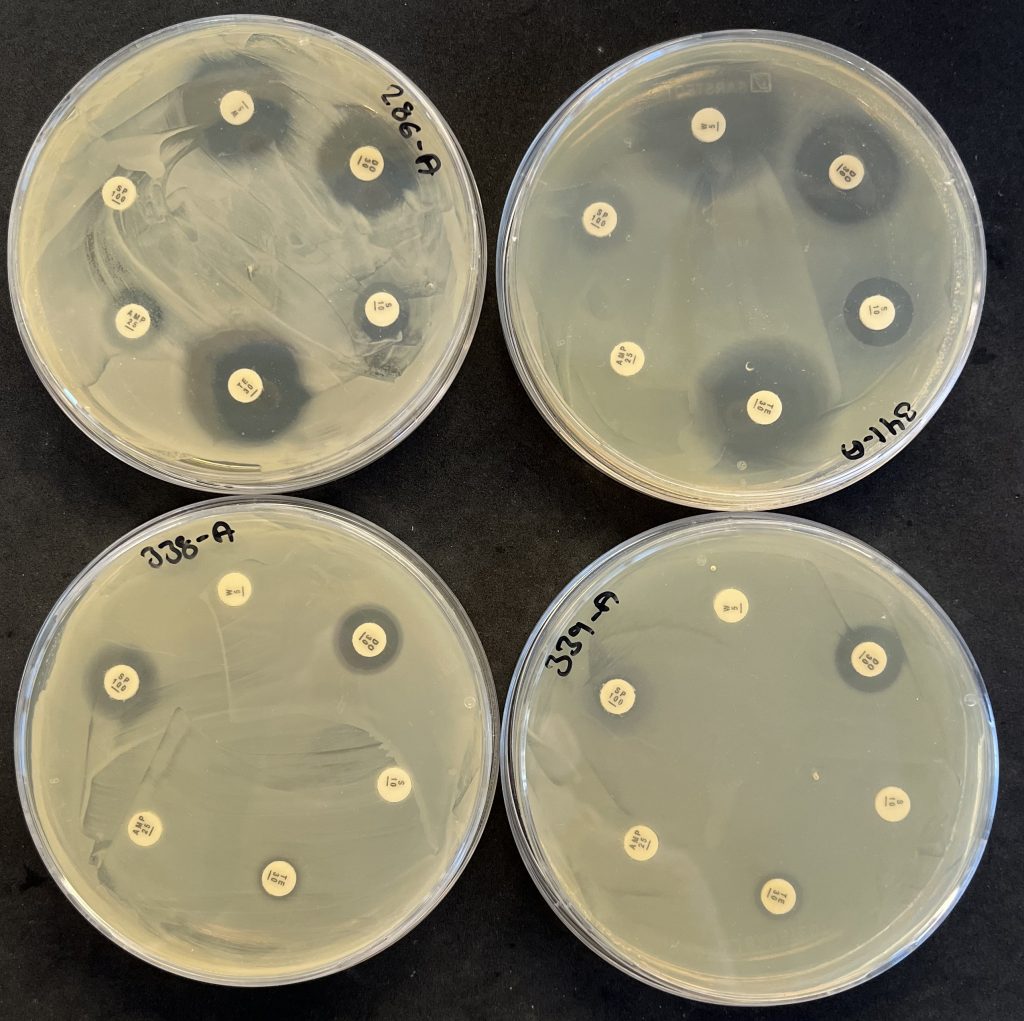
The identified colonies are also tested for multi-resistance by exposing them to 12 different antibiotics. This is done using a clearing-zone method, where agar plates are seeded with a thin layer of the bacterium and small disks loaded with antibiotics are placed on the agar. If the bacterium is susceptible to the antibiotic, it will not grow close to the disk, where the antibiotic concentration is highest, and there will be a clear zone around the disk. If the bacterium is resistant to the antibiotic, it will grow all around the disk, so the narrower the clearing zone, the more resistant is the bacterium. Many of the tested gut bacteria were resistant to several antibiotic types, but none were resistant to all the antibiotics. The below image shows the testing of four petrifilm-bacteria for antibiotic resistance with the clearing zone method. The antibiotics are (clockwise from top): trimethoprim, doxycycline, streptomycin, tetracycline, ampicillin, and spiramycin.
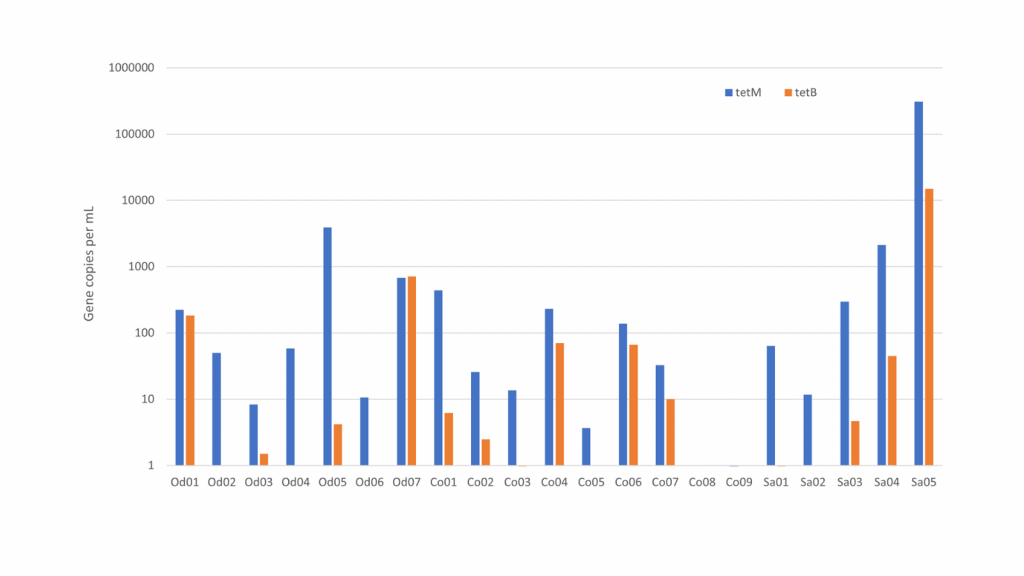
When we get a stormwater sample, we also extract the DNA. The extract is analysed for a range of different antibiotic resistance genes. We use a new method called digital droplet PCR, where we can count the number of copies of each of the resistance genes. With this method, we get a much broader view than with the petrifilm method because we detect many other types of bacteria than the petrifilm enterobacteria and because we also detect bacteria that are hard to grow in the lab.
Results
We found that the load of antibiotic resistance is high in some types of urban stormwater. The highest levels were in sewer overflow and wastewater treatment plant bypass. This is not surprising as both stormwater types are rainwater mixed with domestic sewage. When people are treated with antibiotics, resistant bacteria are enriched in the gut and bladder from where they end up in the wastewater. It was more surprising that we also found elevated levels of antibiotic resistance in surface stormwater in residential areas. These samples presumably were only runoff from streets and roofs with no wastewater, but our counts of fecal E. coli suggest that that there indeed was some degree of wastewater contamination, probably from misconnected pipes.
Next steps
We need to screen more stormwater samples to get precise statistics. We also want to correlate the resistance levels to concentrations of caffeine and theobromine. This is because caffeine and theobromine end up in wastewater when people drink coffee and tea and eat chocolate. Caffeine and theobromine therefore are probably good indicators of domestic wastewater which will help us to discriminate between “pure” surface stormwater and stormwater contaminated with wastewater.
Author: Anders Risbjerg Johnsen (GEUS)
Reference: WHO – World Health Organization, 2022. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022. ISBN 978-92-4-006270-2


